RESEARCH
May 2020
Dongrui Wu and He He
Ministry of Education Key Laboratory of Image Processing and Intelligent Control, School of Artificial Intelligence and Automation, Huazhong University of Science and Technology, Wuhan, China.
Email: drwu@hust.edu.cn, hehe91@hust.edu.cn.
A brain-computer interface (BCI) system [1], [2] acquires the brain signal, decodes it, and then translates it into control commands for external devices, so that a user can interact with his/her surroundings using thoughts directly. Motor imagery (MI) [3] is one of the most frequently used paradigms of BCIs. It is based on the voluntary modulation of the sensorimotor rhythms, which does not need any external stimuli. The imagined movements of different body parts (e.g., hands, feet, and tongue) cause modulations of brain activity in the involved cortical areas.
So, they can be distinguished by decoding such modulations and used to control external devices.
Challenge
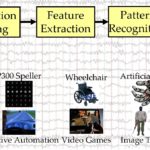
Electroencephalogram (EEG) may be the most popular BCI input signal. The pipeline for decoding EEG signals usually involves signal processing, feature extraction, and pattern recognition, as shown in Fig. 1.
EEG signals are very weak, and easily contaminated by interferences and noise. Moreover, individual differences make it difficult, if not impossible, to build a generic machine learning model optimal for all subjects. Usually a calibration session is needed to collect some subject-specific data for a new subject, which is time-consuming and user-unfriendly. So, reducing this subject-specific calibration is critical to the success of EEG-based BCIs.
State-of-the-art
Researchers have proposed many different approaches [4]–[9] to reduce this calibration effort. One of them is transfer learning [10]. The main idea is to leverage the data from auxiliary subjects (called source subjects or source domains) to improve the learning performance for a new subject (called target subject or target domain).
For example, Zanini et al. [11] proposed a Riemannian alignment (RA) approach to align EEG covariance matrices from different subjects in the Riemannian space. Unfortunately, a Riemannian space classifier is needed after RA, whose computation is usually much more complicated, time-consuming, and less stable than Euclidean space classifiers.
Our Approaches
Inspired by RA, we [12] proposed a Euclidean alignment (EA) approach, which can be used as a preprocessing step before many Euclidean space feature extraction and pattern recognition algorithms.
Both RA and EA assume that the source domains have the same feature space and label space as the target domain, which may not hold in many real-world applications. Recently, we [13] also proposed a label alignment (LA) approach, which can handle situations where the source and target domains have different label spaces. For MI-based BCIs, this means the source subjects and the target subject can perform completely different MI tasks, but the source subjects’ data can still be used to facilitate the calibration for a target subject.
The basic ideas of EA and LA are illustrated in Fig. 2. For illustration, a binary classification case is shown, but both EA and LA can be easily extended to multi-class classification [12], [13]. Initially, the source and target domains scatter far away from each other, due to the domain gap and also the category gap. If we build a classifier on the source domain (indicated by the red dashed line) and apply it directly to the target domain, it may not work at all. EA and LA alleviate this problem by reducing the gaps between the two domains before classification:
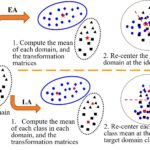
Fig. 2. Illustration of EA and LA. Each EEG trial is represented by its covariance matrix, as a point on a Riemannian manifold. The source domain (blue points) and target domain (black points) represent two different subjects, who have trials from different MI tasks (indicated by different shapes of the points. The shapes in the target domain are only used to help understand our approach, but not to suggest that we need to know all target labels).
1) EA focuses on the domain gap but ignores the category gap completely. It first computes the mean covariance matrix of each domain (indicated by the red stars), from which a transformation matrix of each domain is computed. Using the transformation matrix, EA then re-centers each domain at the identity matrix, and makes the source and target domains overlap with each other, i.e., the domain gap between them is reduced. If we build a classifier in the source domain (the red dashed line) and apply it to the target domain, the classification performance would be improved.
2) LA considers the domain gap and the category gap simultaneously. It first computes the mean covariance matrix of each source domain class (indicated by the red circle and the red diamond) and estimates the mean covariance matrix of each target domain class (indicated by the red triangle and the red square). Then, LA re-centers each source domain class at the corresponding estimated class mean of the target domain. If we build a classifier in the source domain (the red dashed line) and apply it to the target domain, the classification performance would be further improved.
Experimental Validations
We used two publicly available datasets from BCI Competition IV1 to validate EA and LA. The first dataset2 (Dataset 1 [14]) was recorded from seven healthy subjects by 59 EEG channels. Each subject was instructed to perform two classes of MI tasks, selected from three options: left hand, right hand, and feet. The second dataset3 (Dataset 2a) was recorded from nine healthy subjects by 22 EEG channels. Each subject was instructed to perform four classes of MI tasks: left hand, right hand, both feet, and tongue, which were represented by labels 1, 2, 3 and 4, respectively.
In order to intuitively show how EA and LA reduce the distribution discrepancies between the target and source subjects, we project the EEG covariance matrices from the Riemannian manifold into the tangent space, then use the 1D tangent vectors as features to represent the EEG trials. Finally, we use t-stochastic neighbor embedding
(t-SNE) [15], a technique for dimensionality reduction and high-dimensional dataset visualization, to display the
EEG trials (tangent vectors) before and after EA/LA in 2D.
More specifically, we first divide Dataset 2a into two datasets with different label spaces: the source dataset consisted of trials with Labels 1 and 4, and the target dataset with Labels 2 and 3. Then, we pick one subject from the target dataset as the target subject, and the remaining eight subjects from the source dataset as the source subjects. Fig. 3 shows two examples when the first two subjects are used as the target subjects, respectively. The red and black dots are trials of Labels 2 and 3 from the target subject, respectively. The blue and green dots are trials of Labels 1 and 4 from the source subjects, respectively.
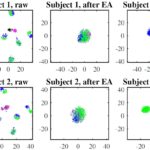
Fig. 3. t-SNE visualization when the first two subjects in Dataset 2a are used as the target subjects, respectively. Red dots: trials of Label 2 from the target subject; black dots: trials of Label 3 from the target subject; blue dots: trials of Label 1 from the source subjects; green dots: trials of Label 4 from the source subjects. The first column shows the trials without alignment, the second column shows the trials after EA, and the third after LA.
Fig. 3 shows that trials from the source subjects (blue and green dots) are scattered far away from those of the target subject (red and black dots), when no alignment is performed. However, the target and source trials overlap with each other after EA, since their centers are now identical. After LA, the target and source trials are further aligned according to their labels. It’s clear that different classes are more distinguishable after LA.
Our extensive classification experiments in [13] demonstrated that:
1) LA only needs as few as one labeled sample from each class of the target subject.
2) LA can be used as a preprocessing step before different feature extraction and classification algorithms.
1http://www.bbci.de/competition/iv/.
2http://www.bbci.de/competition/iv/desc_1.html. 3http://www.bbci.de/competition/iv/desc_2a.pdf.
3) LA can be integrated with other transfer learning approaches to achieve even better classification performance. These conclusions hold true even in the most challenging scenario: the source and target subjects have different feature spaces and also completely different label spaces. For example, we can use the MIs of “left hand” and “right hand” in Dataset I to significantly improve the classification of “feet” and “tongue” MIs in Dataset 2a.
Importance
Our proposed EA is very easy to implement, can be used as a preprocessing step before many Euclidean space feature extraction and pattern recognition algorithms, and can be used for both MI and event related potential classification. Our proposed LA can perform transfer learning between subjects performing completely different MI tasks, which is even more powerful and more flexible than EA. Both EA and LA can significantly help reduce the calibration data requirement for a new subject in EEG-based BCIs, and hence to promote their real-world applications.
Implementations of EA and LA are available at https://github.com/hehe91/EA and https://github.com/hehe91/LA, respectively.
REFERENCES
[1] J. R. Wolpaw, N. Birbaumer, D. J. McFarland, G. Pfurtscheller, and T. M. Vaughan, “Brain-computer interfaces for communication and control,” Clinical Neurophysiology, vol. 113, no. 6, pp. 767–791, 2002.
[2] B. J. Lance, S. E. Kerick, A. J. Ries, K. S. Oie, and K. McDowell, “Brain-computer interface technologies in the coming decades,”
Proc. of the IEEE, vol. 100, no. 3, pp. 1585–1599, 2012.
[3] B. He, B. Baxter, B. J. Edelman, C. C. Cline, and W. W. Ye, “Noninvasive brain-computer interfaces based on sensorimotor rhythms,”
Proc. of the IEEE, vol. 103, no. 6, pp. 907–925, 2015.
[4] V. Jayaram, M. Alamgir, Y. Altun, B. Scholkopf, and M. Grosse-Wentrup, “Transfer learning in brain-computer interfaces,” IEEE Computational Intelligence Magazine, vol. 11, no. 1, pp. 20–31, 2016.
[5] D. Wu, V. J. Lawhern, W. D. Hairston, and B. J. Lance, “Switching EEG headsets made easy: Reducing offline calibration effort using active wighted adaptation regularization,” IEEE Trans. on Neural Systems and Rehabilitation Engineering, vol. 24, no. 11, pp. 1125–1137, 2016.
[6] D. Wu, V. J. Lawhern, S. Gordon, B. J. Lance, and C.-T. Lin, “Driver drowsiness estimation from EEG signals using online weighted adaptation regularization for regression (OwARR),” IEEE Trans. on Fuzzy Systems, vol. 25, no. 6, pp. 1522–1535, 2017.
[7] D. Wu, “Online and offline domain adaptation for reducing BCI calibration effort,” IEEE Trans. on Human-Machine Systems, vol. 47, no. 4, pp. 550–563, 2017.
[8] H. Kang, Y. Nam, and S. Choi, “Composite common spatial pattern for subject-to-subject transfer,” Signal Processing Letters, vol. 16, no. 8, pp. 683–686, 2009.
[9] F. Lotte and C. Guan, “Learning from other subjects helps reducing brain-computer interface calibration time,” in Proc. IEEE Int’l.
Conf. on Acoustics Speech and Signal Processing (ICASSP), Dallas, TX, March 2010.
[10] S. J. Pan and Q. Yang, “A survey on transfer learning,” IEEE Trans. on Knowledge and Data Engineering, vol. 22, no. 10, pp.
1345–1359, 2010.
[11] P. Zanini, M. Congedo, C. Jutten, S. Said, and Y. Berthoumieu, “Transfer learning: a Riemannian geometry framework with applications to brain-computer interfaces,” IEEE Trans. on Biomedical Engineering, vol. 65, no. 5, pp. 1107–1116, 2018.
[12] H. He and D. Wu, “Transfer learning for brain-computer interfaces: A Euclidean space data alignment approach,” IEEE Trans. on Biomedical Engineering, vol. 67, no. 2, pp. 399–410, 2020.
[13] H. He and D. Wu, “Different set domain adaptation for brain-computer interfaces: A label alignment approach,” IEEE Trans. on Neural Systems and Rehabilitation Engineering, 2020, in press. [Online]. Available: https://arxiv.org/abs/1912.01166
[14] B. Blankertz, G. Dornhege, M. Krauledat, K. R. Muller, and G. Curio, “The non-invasive Berlin brain-computer interface: Fast acquisition of effective performance in untrained subjects,” NeuroImage, vol. 37, no. 2, pp. 539–550, 2007.
[15] L. van der Maaten and G. Hinton, “Visualizing data using t-SNE,” Journal of Machine Learning Research, vol. 9, pp. 2579–2605, 2008.
 Dongrui Wu received the BE degree in automatic control from the University of Science and Technology of China in 2003, the ME degree in electrical engineering from the National University of Singapore in 2005, and the PhD degree in electrical engineering from the University of Southern California in 2009. He is now Professor in the School of Artificial Intelligence and Automation, Huazhong University of Science and Technology, Wuhan, China, and Deputy Director of the Key Laboratory of Image Processing and Intelligent Control, Ministry of Education. His research interests include affective computing, brain computer interfaces, computational intelligence, and machine learning. He has more than 140 publications. He is/was an Associate Editor of the IEEE Transactions on Fuzzy Systems (2011-2018), the IEEE Transactions on Human-Machine Systems (2014-), the IEEE Computational Intelligence Magazine (2017-), and the IEEE Transactions on Neural Systems and Rehabilitation Engineering (2019-).
Dongrui Wu received the BE degree in automatic control from the University of Science and Technology of China in 2003, the ME degree in electrical engineering from the National University of Singapore in 2005, and the PhD degree in electrical engineering from the University of Southern California in 2009. He is now Professor in the School of Artificial Intelligence and Automation, Huazhong University of Science and Technology, Wuhan, China, and Deputy Director of the Key Laboratory of Image Processing and Intelligent Control, Ministry of Education. His research interests include affective computing, brain computer interfaces, computational intelligence, and machine learning. He has more than 140 publications. He is/was an Associate Editor of the IEEE Transactions on Fuzzy Systems (2011-2018), the IEEE Transactions on Human-Machine Systems (2014-), the IEEE Computational Intelligence Magazine (2017-), and the IEEE Transactions on Neural Systems and Rehabilitation Engineering (2019-).
 He He received the BE degree in measure and control instrument from the Jiangxi University of Technology in 2013, the ME degree in control science and engineering from the Wuhan University of Technology in 2016. He is now a PhD candidate in the School of Artificial Intelligence and Automation, Huazhong University of Science and Technology, Wuhan, China. His research interests include brain computer interfaces, transfer learning, and machine learning.
He He received the BE degree in measure and control instrument from the Jiangxi University of Technology in 2013, the ME degree in control science and engineering from the Wuhan University of Technology in 2016. He is now a PhD candidate in the School of Artificial Intelligence and Automation, Huazhong University of Science and Technology, Wuhan, China. His research interests include brain computer interfaces, transfer learning, and machine learning.