RESEARCH
May 2020
Abbas Sohrabpour and Bin He
There seem to be two major principles that govern brain function; functional segregation and functional integration [1]. The brain is a highly specialized, and at the same time, a highly integrated organ. Spatially segregated regions are tuned to perform special functions optimally (functional segregation), and at a higher level, multiple regions need to pull resources together, and integrate functions, to perform complex tasks (functional integration). This results in many physiological and mental processes such as cognitive and motor tasks to involve large-scale brain networks functioning together, in harmony. Conversely, many neurological disorders and pathological processes also manifest themselves as malfunctioning networks, such as epilepsy [2]. Logically, the success of studying and, hopefully someday, understanding the brain depends on how well we are equipped with imaging and computational tools suited for analyzing networks. Large-scale brain networks are spatially dispersed and interconnected, following the functional integration principle, yet focal regions on the cortex are co-activated coherently, to fulfill their function. This indicates that extended brain regions will probably be activated at a given interval in the brain. To further complicate matters, these networks are dynamic and change over time in a fast pace, requiring millisecond temporal resolution to discern such variations over time. Basically, these networks are dynamic spatiotemporal processes distributed over the brain. Studying such distributed networks represents a challenge to brain and neuroengineering research.
To study such networks, we need functional imaging modalities that, cover the whole brain and have high spatial and temporal resolution. Electrophysiological recordings, typically, provide high temporal resolution and directly measure brain’s electrical activity as opposed to indirect measures of brain metabolism via hemodynamic responses [3], such as the functional magnetic resonance imaging (fMRI). Among electrophysiological recordings, scalp electromagnetic recordings of brain activity, i.e. the electroencephalography (EEG) and magnetoencephalography (MEG), have two major advantages; firstly, they are noninvasive recordings that pose minimal or no risk for human subjects and, secondly, they also have whole-head coverage. This makes EEG/MEG appropriate tools to study brain networks noninvasively. Scalp electromagnetic recordings such as EEG and MEG have limited spatial resolution, thus, in their raw form lacking sufficient spatial resolution to image brain networks. However, applying proper signal processing and machine learning algorithms, such as electrophysiological source imaging (ESI) techniques, it is possible to delineate underlying sources on the cortical surface with a ~ 5 mm resolution [4]. Recent literature suggests that brain networks can be delineated from surface measurements [5], and even deep sources in the brain can be detected using EEG [6] and MEG [7]. This indicates the important role conventional tools such as EEG and MEG can play in modern neuroimaging and neural engineering applications [8], if proper attention and care is given to developing and applying appropriate and novel computational tools and algorithms.
Brain sources that leave a detectable trace in scalp measurements are extended sources, and it is important to know the location and sizes of these sources to be able to determine the exact region in the cortex responsible for generating a given scalp measurement, which can represent a normal or pathological activity. Most ESI techniques cannot estimate underlying extended brain sources in an objective manner. Recent work has moved towards this direction by employing principles from sparse signal processing literature [9]–[12] or the Bayesian framework [13], [14]. Following our previous development [15], in a recent work [16], we showed that the size and extent of underlying cortical activity can be objectively determined from scalp EEG, using our novel spatiotemporal source imaging algorithm (Figure 1). In our recent publication [16], we studied focal epilepsy patients to determine the epileptogenic tissue from EEG recordings of these patients. We studied 1,027 inter-ictal spikes (sudden huge spikes in recorded EEG of epilepsy patients without any evident clinical symptom) and 86 ictal recordings (paroxysmal high amplitude oscillations observed in epilepsy patients’ EEG recordings during seizures) in these patients to test our hypothesis that the location and extent of underlying epileptogenic tissue can be determined, merely, from high-density scalp EEG (76-electrode montage).
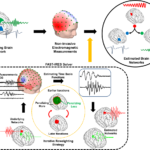
Figure 1. Concept of Network Imaging. Brain networks are distributed spatiotemporal networks and, in our work [16], we modeled them as focally extended sources with coherent temporal activity. The EEG/MEG measurements at scalp, are the net effect of all these underlying sources. Employing blind source separation (BSS) techniques and proper temporal and spatial priors through our FAST-IRES algorithm, we are able to estimate underlying brain networks from EEG measurements. The output of our FAST-IRES model is a spatiotemporal distribution where the location, extent and temporal activity of underlying brain sources can be objectively determined from noninvasive scalp measurements. From [16], licensed under CC BY 4.0.
Epilepsy is a common neurological disorder affecting about 1% of human population with about one third of these patients having uncontrollable seizures despite trying many different anti-convulsant medication [17], [18]. These medically intractable patients often undergo invasive EEG recordings to determine the location and extent of underlying epileptogenic tissue to ultimately undergo surgery and have these troublesome tissues removed. This procedure has become one of the most viable clinical options for these patients [19], if epileptogenic tissues can be accurately localized and removed. As a result our investigation into whether epilepsy networks are discernable from noninvasive scalp measurements can positively affect the quality of care in these patients, potentially supplanting the need for invasive recordings prior to surgery; additionally, as invasive recordings are available in these patients, this information can be used as a testing benchmark and point of reference for our hypothesis; a “ground truth” of sorts.
We used high-density EEG recordings of these patients (prior to surgery) to estimate underlying epilepsy networks and after identifying the epileptogenic tissue using our proposed algorithm, we then compared our results to clinical findings such as the location of intra-cranial EEG (iEEG) electrodes identified as seizure onset (by physicians) and the surgical resection volume (obtained from post-operational MRI). We showed that, on average, localization errors as low as 6 mm and overlaps of 70% (with surgical resection) can be achieved. Additionally, we estimated the time-course of activity for underlying brain sources and demonstrated in a difficult case, where seizure onset zone (SOZ) was deeply located in the hippocampal cortex, that our estimated solution showed a correlation of 0.6 with iEEG recordings from this region; showing that the precise temporal activity of deeply located sources can be estimated from scalp recordings. This allowed us to perform directed connectivity analyses on our estimated solutions to determine the driving nodes of the epilepsy network. Our results indicate that using Granger causality analysis, the estimated epilepsy networks can be investigated systematically to pinpoint the SOZ. We also compared the imaging results from inter-ictal and ictal signals to determine if underlying epileptogenic tissues can be better estimated from one of these signals. We found that ictal imaging results provided more accurate solutions, as some patients can have multiple inconsistent spike types, while ictal recordings, in our cohort, were localized to the affected side (Figure 2).
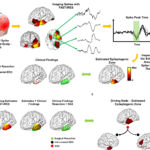
Figure 2. Examples of Imaging Epilepsy Networks in One Patient. The output of FAST-IRES is a spatiotemporal distribution where spatial distributions correspond to a time-course of activity. To determine the epileptogenic tissue, the source signals’ energy is calculated around the spike peak-time and compared to clinical findings for validation (a). An example of inter-ictal imaging (a), ictal imaging (b) and connectivity imaging (c) result for a left temporal epilepsy patient. All results
In summary, our approach indicates that extended large-scale spatiotemporal brain networks can be identified and studied from noninvasive scalp recordings. The location and extent of regions of activity, i.e. network nodes, as well as their temporal evolution can be precisely estimated. This, consequently, enables the researchers to look at the inter-nodal connectivity of these networks, as the temporal activity of nodes can be estimated fairly accurately, which is a requiem for precise computation of inter-nodal connectivity. Our recent work demonstrates that developing novel computational frameworks that incorporate our knowledge of brain networks and structure into their models, is capable of reaching highly accurate results even using well-established measurements such as EEG. It is time to revisit our old acquaintances and reinvent the neuroimaging and neuroengineering field in the light of recent machine learning advancement and novel computational tools.
References
[1] K. J. Friston, “Functional and effective connectivity: a review,” Brain Connect., vol. 1, no. 1, pp. 13–36, 2011. [2] B. C. Bernhardt, L. Bonilha, and D. W. Gross, “Network analysis for a network disorder: The emerging role of graph theory in the study of epilepsy,” Epilepsy Behav., vol. 50, pp. 162–170, Sep. 2015, doi: 10.1016/j.yebeh.2015.06.005. [3] B. J. Edelman, N. Johnson, A. Sohrabpour, S. Tong, N. Thakor, and B. He, “Systems Neuroengineering: Understanding and Interacting with the Brain,” Engineering, vol. 1, no. 3, pp. 292–308, 2015. [4] B. He, A. Sohrabpour, E. Brown, and Z. Liu, “Electrophysiological Source Imaging: A Noninvasive Window to Brain Dynamics,” Annu. Rev. Biomed. Eng., vol. 20, no. 1, pp. 171–196, 2018. [5] A. Sohrabpour, S. Ye, G. A. Worrell, W. Zhang, and B. He, “Noninvasive electromagnetic source imaging and granger causality analysis: An electrophysiological Connectome (eConnectome) approach,” IEEE Trans. Biomed. Eng., vol. 63, no. 12, pp. 2474–2487, 2016. [6] M. Seeber, L.-M. Cantonas, M. Hoevels, T. Sesia, V. Visser-Vandewalle, and C. M. Michel, “Subcortical electrophysiological activity is detectable with high-density EEG source imaging,” Nat. Commun., vol. 10, no. 1, p. 753, Feb. 2019, doi: 10.1038/s41467-019-08725-w. [7] F. Pizzo et al., “Deep brain activities can be detected with magnetoencephalography,” Nat. Commun., vol. 10, no. 1, p. 971, Feb. 2019, doi: 10.1038/s41467-019-08665-5. [8] B. He et al., “Grand challenges in mapping the human brain: NSF workshop report.,” IEEE Trans Biomed Eng., vol. 60, no. 11, pp. 2983–2992, 2013. [9] L. Ding, “Reconstructing cortical current density by exploring sparseness in the transform domain,” Phys. Med. Biol., vol. 54, no. 9, p. 2683, 2009. [10] S. Haufe, V. V. Nikulin, A. Ziehe, K.-R. Müller, and G. Nolte, “Combining sparsity and rotational invariance in EEG/MEG source reconstruction,” NeuroImage, vol. 42, no. 2, pp. 726–738, 2008. [11] A. Gramfort, D. Strohmeier, J. Haueisen, M. S. Hämäläinen, and M. Kowalski, “Time-frequency mixed-norm estimates: Sparse M/EEG imaging with non-stationary source activations,” NeuroImage, vol. 70, pp. 410–422, 2013. [12] P. Krishnaswamy et al., “Sparsity enables estimation of both subcortical and cortical activity from MEG and EEG,” Proc. Natl. Acad. Sci., vol. 114, no. 48, pp. E10465–E10474, Nov. 2017, doi: 10.1073/pnas.1705414114. [13] C. Cai, K. Sekihara, and S. S. Nagarajan, “Hierarchical multiscale Bayesian algorithm for robust MEG/EEG source reconstruction,” NeuroImage, vol. 183, pp. 698–715, Dec. 2018, doi: 10.1016/j.neuroimage.2018.07.056. [14] R. A. Chowdhury et al., “Complex patterns of spatially extended generators of epileptic activity: Comparison of source localization methods cMEM and 4-ExSo-MUSIC on high resolution EEG and MEG data,” NeuroImage, vol. 143, pp. 175–195, Dec. 2016, doi: 10.1016/j.neuroimage.2016.08.044. [15] A. Sohrabpour, Y. Lu, G. Worrell, and B. He, “Imaging brain source extent from EEG/MEG by means of an iteratively reweighted edge sparsity minimization (IRES) strategy,” NeuroImage, vol. 142, pp. 27–42, 2016. [16] A. Sohrabpour, Z. Cai, S. Ye, B. Brinkmann, G. Worrell, and B. He, “Noninvasive electromagnetic source imaging of spatiotemporally distributed epileptogenic brain sources,” Nat. Commun., vol. 11, no. 1, pp. 1–15, Apr. 2020, doi: 10.1038/s41467-020-15781-0. [17] S. L. Moshé, E. Perucca, P. Ryvlin, and T. Tomson, “Epilepsy: new advances,” The Lancet, vol. 385, no. 9971, pp. 884–898, Mar. 2015, doi: 10.1016/S0140-6736(14)60456-6. [18] H. J. Lamberink et al., “Individualised prediction model of seizure recurrence and long-term outcomes after withdrawal of antiepileptic drugs in seizure-free patients: a systematic review and individual participant data meta-analysis,” Lancet Neurol., vol. 16, no. 7, pp. 523–531, Jul. 2017, doi: 10.1016/S1474-4422(17)30114-X. [19] P. Ryvlin, J. H. Cross, and S. Rheims, “Epilepsy surgery in children and adults,” Lancet Neurol., vol. 13, no. 11, pp. 1114–1126, Nov. 2014, doi: 10.1016/S1474-4422(14)70156-5.
Author Biography
 Abbas Sohrabpour studied Electrical Engineering at Sharif University of Technology and later received his PhD in Biomedical Engineering from University of Minnesota, in 2018. He is currently a post-doctoral fellow in Prof. Bin He’s Biomedical Functional Imaging and Neuroengineering Laboratory (BFINL) at Carnegie Mellon University. His current research focuses on investigating signal processing and machine learning techniques in studying biomedical electromagnetic signals such as EEG and MEG, with special interest in studying epilepsy networks.
Abbas Sohrabpour studied Electrical Engineering at Sharif University of Technology and later received his PhD in Biomedical Engineering from University of Minnesota, in 2018. He is currently a post-doctoral fellow in Prof. Bin He’s Biomedical Functional Imaging and Neuroengineering Laboratory (BFINL) at Carnegie Mellon University. His current research focuses on investigating signal processing and machine learning techniques in studying biomedical electromagnetic signals such as EEG and MEG, with special interest in studying epilepsy networks.
 Bin He is Trustee Professor of Biomedical Engineering, Electrical and Computer Engineering, and Neuroscience, and Head of Carnegie Mellon University’s Department of Biomedical Engineering. Dr. He’s research interest mainly lies in systems neuroengineering, including electrophysiological source imaging, noninvasive BCI and neuromodulation. He is the sole editor of textbook “Neural Engineering” with its 3rd edition being published by Springer. His research has been recognized by various awards, including the IEEE William J. Molock Award (2019), the IEEE Biomedical Engineering Award (2017), the IEEE Engineering in Medicine and Biology Society (EMBS) Academic Career Achievement Award (2015), etc. Dr. He served as a Past President of the IEEE EMBS, Editor-in-Chief of IEEE Transactions on Biomedical Engineering.
Bin He is Trustee Professor of Biomedical Engineering, Electrical and Computer Engineering, and Neuroscience, and Head of Carnegie Mellon University’s Department of Biomedical Engineering. Dr. He’s research interest mainly lies in systems neuroengineering, including electrophysiological source imaging, noninvasive BCI and neuromodulation. He is the sole editor of textbook “Neural Engineering” with its 3rd edition being published by Springer. His research has been recognized by various awards, including the IEEE William J. Molock Award (2019), the IEEE Biomedical Engineering Award (2017), the IEEE Engineering in Medicine and Biology Society (EMBS) Academic Career Achievement Award (2015), etc. Dr. He served as a Past President of the IEEE EMBS, Editor-in-Chief of IEEE Transactions on Biomedical Engineering.